| SpectraBase Spectrum ID |
1cPfbFWF10F |
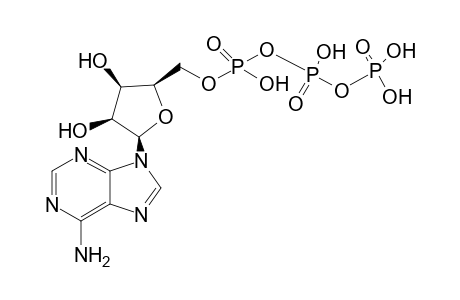
| Name |
ATP |
| Acquisition Mode |
SIMULTANEOUS |
| CAS Registry Number |
10168-83-9
16488-07-6
51569-41-6
56-65-5
71800-44-7
84412-18-0 |
| ChEBI ID |
15422 |
| Comments |
100 mM ATP disodium salt - vendor: Calzyme 45 65 65; Solvent: D2O; Buffers, etc: 50 mM Sodium Phosphate, 500 uM NaAzide; Temperature=298 K, pH=7.4; NMR Reference: 500 uM DSS; Bruker DMX 400MHz
(Data collected by Madison Metabolomics Consortium) |
| Copyright |
Database Compilation Copyright © 2021-2025 John Wiley & Sons, Inc. All Rights Reserved. |
| Data Source |
Madison Metabolomics Consortium |
| Formula |
C10H16N5O13P3 |
| IUPAC Name |
[[[(2R,3R,4R,5R)-5-(6-aminopurin-9-yl)-3,4-dihydroxy-oxolan-2-yl]methoxy-hydroxy-phosphoryl]oxy-hydroxy-phosphoryl]oxyphosphonic acid; [[[(2R,3R,4R,5R)-5-(6-aminopurin-9-yl)-3,4-dihydroxy-tetrahydrofuran-2-yl]methoxy-hydroxy-phosphoryl]oxy-hydroxy-phosphoryl]oxyphosphonic acid |
| InChI |
InChI=1S/C10H16N5O13P3/c11-8-5-9(13-2-12-8)15(3-14-5)10-7(17)6(16)4(26-10)1-25-30(21,22)28-31(23,24)27-29(18,19)20/h2-4,6-7,10,16-17H,1H2,(H,21,22)(H,23,24)(H2,11,12,13)(H2,18,19,20)/t4-,6+,7+,10-/m1/s1 |
| InChIKey |
ZKHQWZAMYRWXGA-XJCFNFQFSA-N |
| KEGG Compound ID |
C00002 |
| KEGG Pathways |
PATH: map00190 Oxidative phosphorylation
PATH: map00195 Photosynthesis
PATH: map00230 Purine metabolism
PATH: map00231 Puromycin biosynthesis
PATH: map04020 Calcium signaling pathway
PATH: map04080 Neuroactive ligand-receptor interaction
PATH: map04930 Type II diabetes mellitus
PATH: map05110 Cholera û Infection |
| PubChem Compound ID |
5957 |
| SMILES |
C1=NC2=C(C(=N1)N)N=CN2C3C(C(C(O3)COP(=O)(O)OP(=O)(O)OP(=O)(O)O)O)O |
| Source File Reference |
bmse000006 |